

|
PhD ProjectThe primary aim for this project was to create a spatio-temporal data set of a growing Arabidopsis leaf. Currently in the lab at the John Innes Centre we have access to an Optical Projection Tomography scanner. Using the scanner it's possible to produce volumetric scans of plant organs. 
For this project I am using OPT to build a data set of Arabidopsis leaves captured at different developmental stages. I created a volumetric rendering application to allow for the visualisation of such volume images. The application is implemented in C++/Qt/OpenGL/GLSL and was named QtVolViewer. 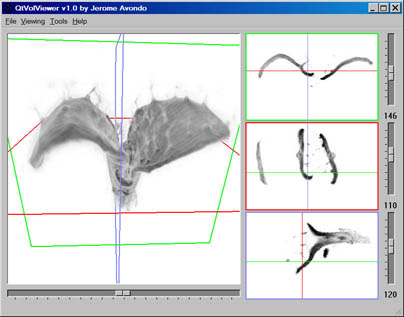
The idea of this project is to use the trichome cells, hairs found on the leaf surface, as markers for our leaf. Surface classification algorithms have been implemented to test different techniques to indentify and extract these hairs from our volumetric data. Below is a successfully segmented lamina and identified trichomes. 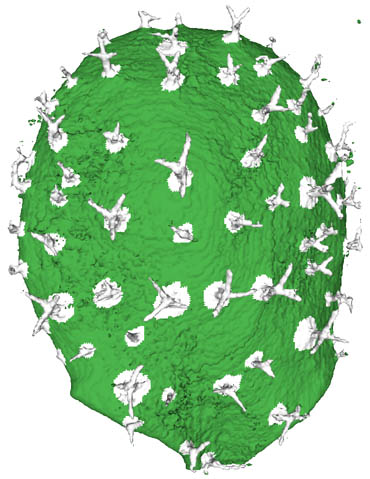

Once the trichomes are found, further processing is applied to extract the orientation and base for each individual trichome. This is achieved using principle component analysis and k-means clustering. 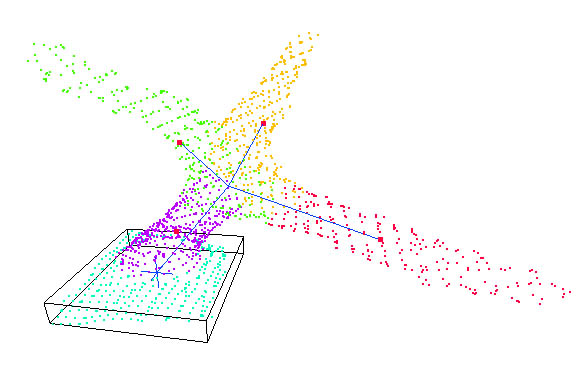
I am currently at the stage that the tools have been successfully implemented and tested. I am now trying to generate a large data set of Arabidopsis leaves. Future work is to investigate what kind of growth parameters can be extracted from this data set. |